Typing of S. epidermidis samples (MD P.B.)
gene_x 0 like s 1194 view s
Tags: bioinformatics, pipeline, DNA-seq
Characterization_of_the_virulence_agr_typing_and_antimicrobial_resistance_profile_of_Staphylococcus_aureus_strains.pdf
-
Goal of data analyses
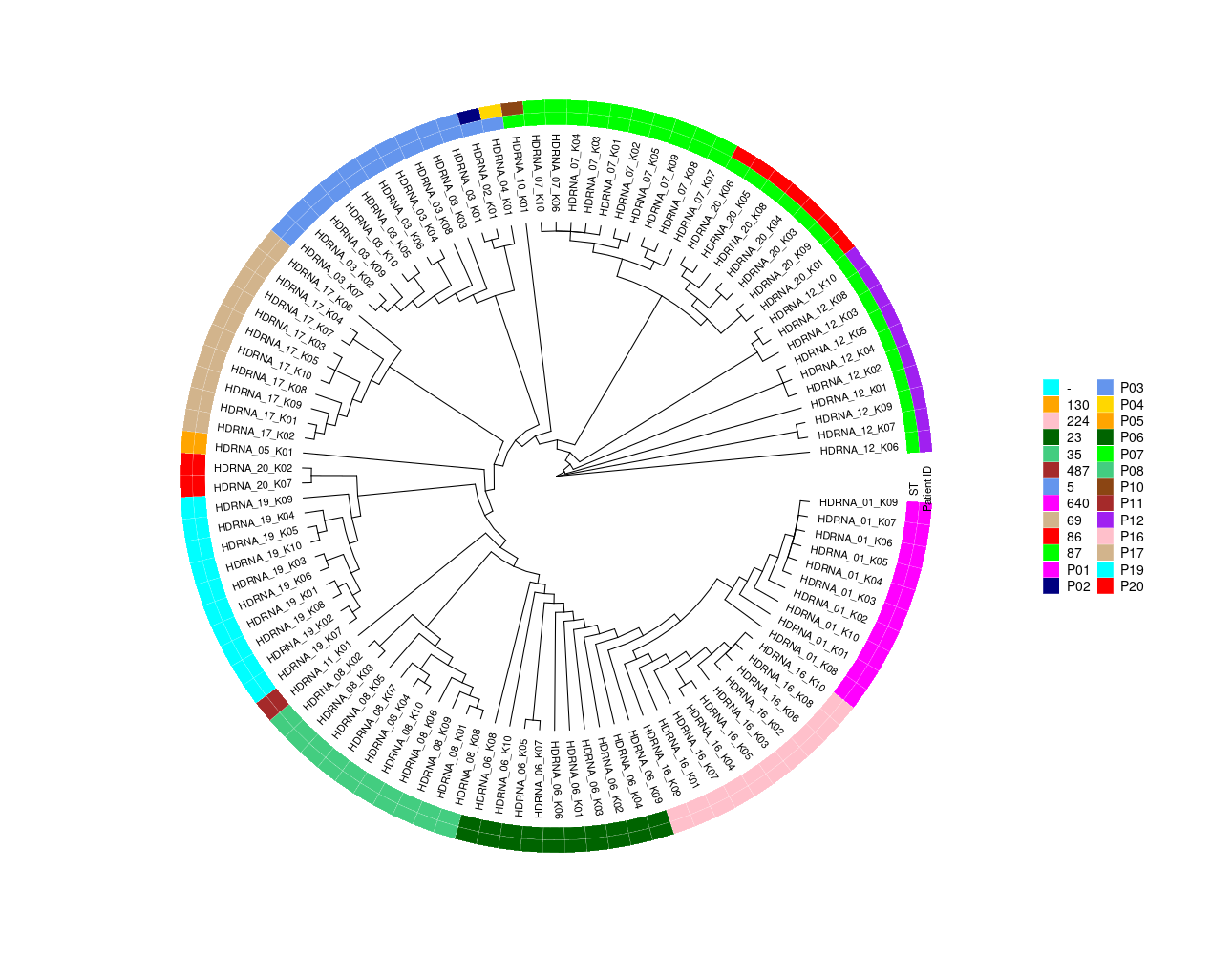
S epidermidis genomes 1. Assemble closed genomes from HDRNA 1, 3, 6, 7, 8, 12, 16, 17, 19, 20 (short read + long read) TODO: make a table similar to the paper Characterization of the virulence, agr typing and antimicrobial resistance profile of Staphylococcus aureus strains isolated from food handlers in Brazil Table 2, and draw a tree+heatmaps Figure! Based on closed genomes: - Sequence type, - goeBURST analysis (-->performed is geoBURST TLV-Analysis, see point 2), - SCCmec type (https://www.sccmec.org/index.php/en/method-to-identify-sccmmcc-smn-en/review-smn-en) https://cge.cbs.dtu.dk/services/SCCmecFinder/ SCCmec typing: https://www.genomicepidemiology.org/ --> https://cge.food.dtu.dk/services/SCCmecFinder/ SCCmecFinder 1.2 SCCmecFinder identifies SCCmec elements in sequenced S. aureus isolates. The SCCmec element is the defining feature of methicillin-resistant S. aureus isolates, and encodes the single determinant for methicillin resistant, the mecA gene. IMPORTANT! SCCmec typing is only available for SCCmec type I-XI and subtyping is currently only available for SCCmec type IV and V IMPORTANT! mec gene complex C1 and C2 might produce errors. - agr typing: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187671/ (see point 3) - presence phage HH1, SPbeta-like phage, phage related island [referring to A´s paper] (see point 5), - absence/presence matrix for selected genes [see attached ppt, see results in Gene_List.pptx] (see point 6) gyrB, fumC, , icd, apsS, sigB, sarA, , , , , , sdrG(-17), sdrH, ebh, ebp (ebpS), , , , dltA, , lipA, , , , , , , --> draw a circle heatmaps including all data with a very big figure and table. 2. according to assembled genomes describe within host diversity per patient (i.e. compare isolates 2 – 10 to isolate 1). python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_01_K01_conservative_23197.current.gb HDRNA_01_K01_CP133676.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_03_K01_bold_bandage_26831.current.gb HDRNA_03_K01_CP133677.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_06_K01_conservative_27645.current.gb HDRNA_06_K01_CP133678-CP133679.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_07_K01_conservative_27169.current.gb HDRNA_07_K01_CP133680-CP133681.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_08_K01_conservative_32455.current.gb HDRNA_08_K01_CP133682-CP133683.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_12_K01_bold_37467.current.gb HDRNA_12_K01_CP133684-CP133687.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_16_K01_conservative_37834.current.gb HDRNA_16_K01_CP133688-CP133692.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_17_K01_conservative_37288.current.gb HDRNA_17_K01_CP133693-CP133695.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_19_K01_bold_37377.current.gb HDRNA_19_K01_CP133696-CP133699.xlsx python3 /home/jhuang/Scripts/gb_to_excel.py ./gbks/HDRNA_20_K01_conservative_43457.current.gb HDRNA_01_K01_CP133700-CP133701.xlsx * SNP INDEL: snippy+spandx --> get the complete list of SNP+INDEL for each isolate group!!! (see point 4) * gene absence/presence: # A: prepare prokka_HDRNA_01 .. prokka_HDRNA_20 from prokka_remaining # B: #https://github.com/jorvis/biocode/blob/master/gff/convert_genbank_to_gff3.py sudo apt-get install -y python3 python3-pip zlib1g-dev libblas-dev liblapack-dev libxml2-dev pip3 install biocode convert_genbank_to_gff3.py -i HDRNA_01_K01_conservative_23197.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_01/HDRNA_01_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_03_K01_bold_bandage_26831.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_03/HDRNA_03_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_06_K01_conservative_27645.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_06/HDRNA_06_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_07_K01_conservative_27169.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_07/HDRNA_07_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_08_K01_conservative_32455.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_08/HDRNA_08_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_12_K01_bold_37467.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_12/HDRNA_12_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_16_K01_conservative_37834.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_16/HDRNA_16_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_17_K01_conservative_37288.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_17/HDRNA_17_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_19_K01_bold_37377.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_19/HDRNA_19_K01.gff --with_fasta convert_genbank_to_gff3.py -i HDRNA_20_K01_conservative_43457.current.gb -o ~/DATA/Data_PaulBongarts_S.epidermidis_HDRNA/Data_Holger_S.epidermidis_short/prokka_HDRNA_20/HDRNA_20_K01.gff --with_fasta # C: check gff.files if containing the repeated gene names using check_duplicate_cds.py, remove manually, recheck the removed one. python3 ~/Scripts/check_duplicate_cds.py HDRNA_01_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_03_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_06_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_07_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_08_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_12_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_16_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_17_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_19_K01.gff python3 ~/Scripts/check_duplicate_cds.py HDRNA_20_K01.gff Found duplicates for the following CDS IDs: RE430_09810.mRNA.0.CDS.1 RE430_08580.mRNA.0.CDS.1 RE430_09730.mRNA.0.CDS.1 RGR13_00575.mRNA.0.CDS.1 RGR10_09570.mRNA.0.CDS.1 RGR06_10025.mRNA.0.CDS.1 RGR06_12305.mRNA.0.CDS.1 #xxxx RGR12_09490.mRNA.0.CDS.1 RGR12_00425.mRNA.0.CDS.1 RGR12_11570.mRNA.0.CDS.1 RGR09_12270.mRNA.0.CDS.1 RGR09_09635.mRNA.0.CDS.1 RGR09_01280.mRNA.0.CDS.1 RGR08_01845.mRNA.0.CDS.1 RGR08_13135.mRNA.0.CDS.1 RGR08_10360.mRNA.0.CDS.1 RGR08_09845.mRNA.0.CDS.1 RGR14_00635.mRNA.0.CDS.1 RGR14_00355.mRNA.0.CDS.1 RGR14_09870.mRNA.0.CDS.1 RGR14_01550.mRNA.0.CDS.1 RGR07_09075.mRNA.0.CDS.1 RGR07_00190.mRNA.0.CDS.1 RGR07_11485.mRNA.0.CDS.1 RGR07_00090.mRNA.0.CDS.1 RGR07_11545.mRNA.0.CDS.1 RGR07_01915.mRNA.0.CDS.1 RGR11_01280.mRNA.0.CDS.1 RGR11_09700.mRNA.0.CDS.1 rsync -a -P prokka_HDRNA_01/HDRNA_01_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_01/ rsync -a -P prokka_HDRNA_03/HDRNA_03_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_03/ rsync -a -P prokka_HDRNA_06/HDRNA_06_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_06/ rsync -a -P prokka_HDRNA_07/HDRNA_07_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_07/ rsync -a -P prokka_HDRNA_08/HDRNA_08_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_08/ rsync -a -P prokka_HDRNA_12/HDRNA_12_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_12/ rsync -a -P prokka_HDRNA_16/HDRNA_16_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_16/ rsync -a -P prokka_HDRNA_17/HDRNA_17_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_17/ rsync -a -P prokka_HDRNA_19/HDRNA_19_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_19/ rsync -a -P prokka_HDRNA_20/HDRNA_20_K01.gff jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_20/ cd prokka_HDRNA_01 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_01_K01.gff ./HDRNA_01_K02/HDRNA_01_K02.gff ./HDRNA_01_K03/HDRNA_01_K03.gff ./HDRNA_01_K04/HDRNA_01_K04.gff ./HDRNA_01_K05/HDRNA_01_K05.gff ./HDRNA_01_K06/HDRNA_01_K06.gff ./HDRNA_01_K07/HDRNA_01_K07.gff ./HDRNA_01_K08/HDRNA_01_K08.gff ./HDRNA_01_K09/HDRNA_01_K09.gff ./HDRNA_01_K10/HDRNA_01_K10.gff cd ../prokka_HDRNA_03 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_03_K01.gff ./HDRNA_03_K02/HDRNA_03_K02.gff ./HDRNA_03_K03/HDRNA_03_K03.gff ./HDRNA_03_K04/HDRNA_03_K04.gff ./HDRNA_03_K05/HDRNA_03_K05.gff ./HDRNA_03_K06/HDRNA_03_K06.gff ./HDRNA_03_K07/HDRNA_03_K07.gff ./HDRNA_03_K08/HDRNA_03_K08.gff ./HDRNA_03_K09/HDRNA_03_K09.gff ./HDRNA_03_K10/HDRNA_03_K10.gff cd ../prokka_HDRNA_06 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_06_K01.gff ./HDRNA_06_K02/HDRNA_06_K02.gff ./HDRNA_06_K03/HDRNA_06_K03.gff ./HDRNA_06_K04/HDRNA_06_K04.gff ./HDRNA_06_K05/HDRNA_06_K05.gff ./HDRNA_06_K06/HDRNA_06_K06.gff ./HDRNA_06_K07/HDRNA_06_K07.gff ./HDRNA_06_K08/HDRNA_06_K08.gff ./HDRNA_06_K09/HDRNA_06_K09.gff ./HDRNA_06_K10/HDRNA_06_K10.gff cd ../prokka_HDRNA_07 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_07_K01.gff ./HDRNA_07_K02/HDRNA_07_K02.gff ./HDRNA_07_K03/HDRNA_07_K03.gff ./HDRNA_07_K04/HDRNA_07_K04.gff ./HDRNA_07_K05/HDRNA_07_K05.gff ./HDRNA_07_K06/HDRNA_07_K06.gff ./HDRNA_07_K07/HDRNA_07_K07.gff ./HDRNA_07_K08/HDRNA_07_K08.gff ./HDRNA_07_K09/HDRNA_07_K09.gff ./HDRNA_07_K10/HDRNA_07_K10.gff cd ../prokka_HDRNA_08 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_08_K01.gff ./HDRNA_08_K02/HDRNA_08_K02.gff ./HDRNA_08_K03/HDRNA_08_K03.gff ./HDRNA_08_K04/HDRNA_08_K04.gff ./HDRNA_08_K05/HDRNA_08_K05.gff ./HDRNA_08_K06/HDRNA_08_K06.gff ./HDRNA_08_K07/HDRNA_08_K07.gff ./HDRNA_08_K08/HDRNA_08_K08.gff ./HDRNA_08_K09/HDRNA_08_K09.gff ./HDRNA_08_K10/HDRNA_08_K10.gff cd ../prokka_HDRNA_12 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_12_K01.gff ./HDRNA_12_K02/HDRNA_12_K02.gff ./HDRNA_12_K03/HDRNA_12_K03.gff ./HDRNA_12_K04/HDRNA_12_K04.gff ./HDRNA_12_K05/HDRNA_12_K05.gff ./HDRNA_12_K06/HDRNA_12_K06.gff ./HDRNA_12_K07/HDRNA_12_K07.gff ./HDRNA_12_K08/HDRNA_12_K08.gff ./HDRNA_12_K09/HDRNA_12_K09.gff ./HDRNA_12_K10/HDRNA_12_K10.gff cd ../prokka_HDRNA_16 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_16_K01.gff ./HDRNA_16_K02/HDRNA_16_K02.gff ./HDRNA_16_K03/HDRNA_16_K03.gff ./HDRNA_16_K04/HDRNA_16_K04.gff ./HDRNA_16_K05/HDRNA_16_K05.gff ./HDRNA_16_K06/HDRNA_16_K06.gff ./HDRNA_16_K07/HDRNA_16_K07.gff ./HDRNA_16_K08/HDRNA_16_K08.gff ./HDRNA_16_K09/HDRNA_16_K09.gff ./HDRNA_16_K10/HDRNA_16_K10.gff cd ../prokka_HDRNA_17 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_17_K01.gff ./HDRNA_17_K02/HDRNA_17_K02.gff ./HDRNA_17_K03/HDRNA_17_K03.gff ./HDRNA_17_K04/HDRNA_17_K04.gff ./HDRNA_17_K05/HDRNA_17_K05.gff ./HDRNA_17_K06/HDRNA_17_K06.gff ./HDRNA_17_K07/HDRNA_17_K07.gff ./HDRNA_17_K08/HDRNA_17_K08.gff ./HDRNA_17_K09/HDRNA_17_K09.gff ./HDRNA_17_K10/HDRNA_17_K10.gff cd ../prokka_HDRNA_19 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_19_K01.gff ./HDRNA_19_K02/HDRNA_19_K02.gff ./HDRNA_19_K03/HDRNA_19_K03.gff ./HDRNA_19_K04/HDRNA_19_K04.gff ./HDRNA_19_K05/HDRNA_19_K05.gff ./HDRNA_19_K06/HDRNA_19_K06.gff ./HDRNA_19_K07/HDRNA_19_K07.gff ./HDRNA_19_K08/HDRNA_19_K08.gff ./HDRNA_19_K09/HDRNA_19_K09.gff ./HDRNA_19_K10/HDRNA_19_K10.gff cd ../prokka_HDRNA_20 roary -p 5 -f ./roary -i 95 -cd 99 -s -e -n -v ./HDRNA_20_K01.gff ./HDRNA_20_K02/HDRNA_20_K02.gff ./HDRNA_20_K03/HDRNA_20_K03.gff ./HDRNA_20_K04/HDRNA_20_K04.gff ./HDRNA_20_K05/HDRNA_20_K05.gff ./HDRNA_20_K06/HDRNA_20_K06.gff ./HDRNA_20_K07/HDRNA_20_K07.gff ./HDRNA_20_K08/HDRNA_20_K08.gff ./HDRNA_20_K09/HDRNA_20_K09.gff cd .. rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_01/roary prokka_HDRNA_01 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_03/roary prokka_HDRNA_03 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_06/roary prokka_HDRNA_06 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_07/roary prokka_HDRNA_07 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_08/roary prokka_HDRNA_08 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_12/roary prokka_HDRNA_12 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_16/roary prokka_HDRNA_16 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_17/roary prokka_HDRNA_17 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_19/roary prokka_HDRNA_19 rsync -a -P jhuang@hamm:~/DATA/Data_Holger_S.epidermidis_short/prokka_HDRNA_20/roary prokka_HDRNA_20 cp prokka_HDRNA_01/roary/gene_presence_absence.csv gene_presence_absence__HDRNA_01.csv cp prokka_HDRNA_03/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_03.csv cp prokka_HDRNA_06/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_06.csv cp prokka_HDRNA_07/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_07.csv cp prokka_HDRNA_08/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_08.csv cp prokka_HDRNA_12/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_12.csv cp prokka_HDRNA_16/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_16.csv cp prokka_HDRNA_17/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_17.csv cp prokka_HDRNA_19/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_19.csv cp prokka_HDRNA_20/roary/gene_presence_absence.csv gene_presence_absence_HDRNA_20.csv #Wenn man will open the files mit libreoffice, needs "," --> "|"; "\n"-->\n; the first and last " in the text; in the kate, then open file with libreoffice with delimiter '|'. sed -i 's/\.mRNA\.0\.CDS\.1//g' gene_presence_absence__HDRNA_01.csv group_10||Y_phosphoryl: pyrimidine-nucleoside phosphorylase|10|11|1.11|1|416||||362|1301|1165|RE430_03860|HDRNA_01_K02_02111|HDRNA_01_K03_02181|HDRNA_01_K04_02110|HDRNA_01_K05_02181|HDRNA_01_K06_02156|HDRNA_01_K07_02196 HDRNA_01_K07_02197|HDRNA_01_K08_01754|HDRNA_01_K09_02226|HDRNA_01_K10_02107 group_10||Y_phosphoryl: pyrimidine-nucleoside phosphorylase|10|11|1.11|1|416||||362|1301|1165|RE430_03860.mRNA.0.CDS.1|HDRNA_01_K02_02111|HDRNA_01_K03_02181|HDRNA_01_K04_02110|HDRNA_01_K05_02181|HDRNA_01_K06_02156|HDRNA_01_K07_02196 HDRNA_01_K07_02197|HDRNA_01_K08_01754|HDRNA_01_K09_02226|HDRNA_01_K10_02107 for file in gene_presence_absence_HDRNA_*; do sed -i 's/\.mRNA\.0\.CDS\.1//g' "$file" done ~/Tools/csv2xls-0.4/csv_to_xls.py gene_presence_absence__HDRNA_01.csv gene_presence_absence_HDRNA_03.csv gene_presence_absence_HDRNA_06.csv gene_presence_absence_HDRNA_07.csv gene_presence_absence_HDRNA_08.csv gene_presence_absence_HDRNA_12.csv gene_presence_absence_HDRNA_16.csv gene_presence_absence_HDRNA_17.csv gene_presence_absence_HDRNA_19.csv gene_presence_absence_HDRNA_20.csv -d'|' -o gene_presence_absence.xls * genomic rearrangements (e.g. SCCmec deletions, ACME deletions, agr insertions) TODOs using Easyfig! #Staphylococcal Cassette Chromosome mec #arginine catabolic mobile element (ACME) #Prevalence and genetic diversity of arginine catabolic mobile element (ACME) in clinical isolates of coagulase-negative staphylococci: identification of ACME type I variants in Staphylococcus epidermidis. Fig. 1. A schematic drawing of genetic structures of ACME (a region from the arc to opp3 cluster, or corresponding genetic components) among the three DI subtypes (DI.1, DI.2, and DI.3: strains CNS266, CNS115, and CNS149, respectively), type I (strain USA300-FPR3757, accession number CP000255), type II (strain ATCC12228, accession number AE015929) and type DII (strain M08/0126, accession number FR753166). Putative ORFs of genes are represented by arrows colored with green (arc cluster), red (opp3 cluster), blue (a region between the arc and opp3 clusters in ACME I), or dark blue (genes in ACME II). The regions in light pink including the arc cluster indicate genetically identical areas to both ATCC12228 and USA300-FPR3757. The regions with light blue are identical to only ATCC12228, while those with light orange to USA300-FPR3757. White space regions between argR and SAUSA300_0072 show no sequence homology either to ATCC12228 or to USA300_FPR3757; however, these regions show 91–99% nucleotide identity among the three ACME subtypes. Regions colored with dark orange in the three ACME DI subtypes show=98% nucleotide sequence identity to each other. Regions colored with grey (type DI.1), purple or cyan (type DI.3) do not show high nucleotide identity (<98%) to cognate genes in other ACME types (Table S2.2). Positions of primers used for PCR profile (Tables 1 and 4) are shown with arrowheads under ACME I sequence. Collapse #Smash++: https://academic.oup.com/gigascience/article/9/5/giaa048/5841055 #https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7597632/ Artemis Comparison Tool (ACT): Allows for the visual comparison of genomes and can be used to investigate the presence or absence of genomic regions (such as SCCmec or ACME) and other structural variations. #https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009304 #https://journals.plos.org/plospathogens/article/figure?id=10.1371/journal.ppat.1009304.g010 The image you uploaded appears to be a schematic representation of genomic rearrangements, typically found in scientific publications or reports. These kinds of images are often created using bioinformatics visualization tools or general graphic design software. While it's not possible to determine the exact software used to create this specific image without more context, I can suggest several tools commonly used for such purposes: 3. Epidome: (using R?) Create bar plot showing ST distribution in noses from patients HDRNA 1, 3, 6, --> 7, 8, 12, 16, 17, 19, 20 <-- (Easy using R) Create table showing presence / absence of STs / per patient NOTE: Epidome data have not been processed due to potential missing of the data; At first sending the results without Epidome results, ask again where is the epidome data for the 10 patients? -
generate goeBURST
5,1,1,1,2,2,1,1 87,7,1,1,2,2,1,1 35,2,1,2,2,4,1,1 69,1,18,6,2,2,1,1 23,7,1,2,1,3,3,1 224,19,16,19,6,3,19,10 640,28,3,13,5,8,9,11 '-',1,13,2,1,2,1,29 in goeBURST-1.2.1.jar, I have gelesen "Edge level to define group SLV DLV TLV". Wie kann ich generate SLV, DLV and TLV files? The input you've provided seems to represent allelic profiles or sequence types (STs) used in microbial typing, particularly in methods like Multilocus Sequence Typing (MLST). When you input these profiles into software like goeBURST (implemented in a tool like PHYLOViZ), the software uses these profiles to construct a phylogenetic network. The network shows relationships between different strains or isolates based on their allelic similarity. In goeBURST, groups are defined based on their allelic differences: SLV (Single-Locus Variants): Strains or STs differing by only one locus. DLV (Double-Locus Variants): Strains or STs differing by two loci. TLV (Triple-Locus Variants): Strains or STs differing by three loci. -
agr typing S. epidermidis
> Comparison of the amino acid sequences of a region of the N-terminus of AgrC (A), and AgrB (B) of S. epidermidis (S.e.), S. aureus (S.a.) and S. lugdunensis (S.l.). > https://academic.oup.com/femsle/article/163/1/1/625220: Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5064449/ https://brieflands.com/articles/archcid-62833#4.-Results https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1282 Staphylococcus epidermidis Agr Operon https://academic.oup.com/femspd/article/51/1/220/501159 #pAgr (S. epidermidis agr operon promoter) https://parts.igem.org/Part:BBa_K212003# - An overall homology of 68% was found between the agr locus from S. epidermidis and S. aureus. - The agr locus from S. epidermidis was organized similar to those from S. aureus and S. lugdunensis. - The putative RNAII molecule contains four open reading frames, agrA, B, C and D. AgrA was a response regulator. - AgrB showed homology with transducer and translocase molecules. - AgrC is expected to act as a histidine protein kinase in which a leucine zipper is present. - AgrD is presumably processed into an autoinducer peptide. - For a long time, Staphylococcus epidermidis, as a member of the coagulase-negative staphylococci, was considered as part of the physiological skin flora of the human being with no pathogenic significance. - Today, we know that S. epidermidis is one of the most prevalent causes for implant-associated and nosocomial infections. - We performed pheno- and genotypic analysis (ica, IS256, SCCmec types, agr groups) of biofilm formation in 200 isolates. - Fifty percent were genetically ica-positive and produced biofilm. - Among all studied isolates, agr II and III and SCCmec type I were the most prevalent, whereas within the selected multi-resistant isolates (29%), agr I and III and SCCmec type II dominated. - SCCmec type I and mecA-negative S. epidermidis isolates were associated with agr II. - The majority of the blood culture and biopsy isolates were assigned to agr III and SCCmec type I, whereas agr II was predominantly detected in mecA-negative S. epidermidis isolated from catheter and implant materials. - MLST analysis revealed the major clonal lineages of ST2, ST5, ST10, and ST242 (total 13 STs). - ST2 isolates from blood cultures were icaA/D-positive and harbored SCCmec types II and III and IS256, whereas the icaA/D- and IS256-positive ST23 isolates were assigned to SCCmec types I and IV. # -- OPTION 1 using AgrVATE gp1-4-operon_ref.fasta (failed since the database contains only agr from Staphylococcus aureus) -- > Species-Wide Phylogenomics of the Staphylococcus aureus Agr Operon Revealed Convergent Evolution of Frameshift Mutations makeblastdb -in HDRNA_K01.fna -dbtype nucl -perc_identity 90 -qcov_hsp_perc 90 blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query /home/jhuang/Tools/AgrVATE/agrvate_databases/references/gp1-4-operon_ref.fasta -evalue 0.1 -num_threads 15 -outfmt "6 sseqid qseqid evalue pident sstart send" -strand both -max_target_seqs 1 blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query gp1-4-operon_ref.fasta -evalue 0.1 -num_threads 15 -outfmt "6 sseqid qseqid evalue pident sstart send" -strand both > HDRNA_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query /home/jhuang/Tools/AgrVATE/agrvate_databases/references/gp1-4-operon_ref.fasta -evalue 0.1 -num_threads 15 -outfmt "6 sseqid qseqid evalue pident sstart send" -strand both > HDRNA_03.blastn blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query /home/jhuang/Tools/AgrVATE/agrvate_databases/gp1234_all_motifs.fna -evalue 0.1 -num_threads 15 -outfmt "6 sseqid qseqid evalue pident sstart send" -strand both > HDRNA_01_.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query /home/jhuang/Tools/AgrVATE/agrvate_databases/gp1234_all_motifs.fna -evalue 0.1 -num_threads 15 -outfmt "6 sseqid qseqid evalue pident sstart send" -strand both > HDRNA_03_.blastn # -- OPTION 2 using agr-1-3_Se.fasta (failed!) -- > High Genetic Variablity of the agr Locus in Staphylococcus Species >gi|3320006|emb|Z49220.1| Staphylococcus epidermidis hld and agr[A,B,C,D] genes >gi|18251022|gb|AF346724.1| Staphylococcus epidermidis strain N910160 AgrB (agrB) gene, partial cds; AgrD (agrD) gene, complete cds; and AgrC (agrC) gene, partial cds >gi|18251026|gb|AF346725.1| Staphylococcus epidermidis strain N910191 AgrB (agrB) gene, partial cds; AgrD (agrD) gene, complete cds; and AgrC (agrC) gene, partial cds blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query agr-1-3_Se.fasta -evalue 0.1 -num_threads 15 -outfmt 6 -strand both > HDRNA_01__.blastn # -- OPTION 3 using agrD_I-III.fasta (successful!) -- > Staphylococcus epidermidis agr Quorum-Sensing System: Signal Identification, Cross Talk, and Importance in Colonization > https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4187671/ > https://www.researchgate.net/publication/264391883 tblastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_01.tblastn tblastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_03.tblastn tblastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_06.tblastn tblastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_07.tblastn tblastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_08.tblastn tblastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_12.tblastn tblastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_16.tblastn tblastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_17.tblastn tblastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_19.tblastn tblastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query agrD_I-III.fasta -evalue 0.1 -num_threads 15 > HDRNA_20.tblastn II? II I II I II II II III II >AgrD_I MENIFNLFIKFFTTILEFIGTVAGDSVCASYFDEPEVPEELTKLYE >AgrD_II MNLLGGLLLKIFSNFMAVIGNASKYNPCSNYLDEPQVPEELTKLDE >AgrD_III MNLLGGLLLKLFSNFMAVIGNAAKYNPCASYLDEPQVPEELTKLDE -
Variant (SNP+INDEL) calling
Input files: HDRNA_01_K01_conservative_23197.current.gb HDRNA_01_K01_conservative_23197.current.gb:LOCUS CP133676 2502964 bp DNA circular BCT 05-SEP-2023 HDRNA_03_K01_bold_bandage_26831.current.gb:LOCUS CP133677 2590275 bp DNA circular BCT 05-SEP-2023 HDRNA_06_K01_conservative_27645.current.gb:LOCUS CP133678 2465260 bp DNA circular BCT 05-SEP-2023 HDRNA_06_K01_conservative_27645.current.gb:LOCUS CP133679 19348 bp DNA circular BCT 05-SEP-2023 HDRNA_07_K01_conservative_27169.current.gb:LOCUS CP133680 2544074 bp DNA circular BCT 05-SEP-2023 HDRNA_07_K01_conservative_27169.current.gb:LOCUS CP133681 2241 bp DNA circular BCT 05-SEP-2023 HDRNA_08_K01_conservative_32455.current.gb:LOCUS CP133682 2425353 bp DNA circular BCT 05-SEP-2023 HDRNA_08_K01_conservative_32455.current.gb:LOCUS CP133683 6358 bp DNA circular BCT 05-SEP-2023 HDRNA_12_K01_bold_37467.current.gb:LOCUS CP133684 2490139 bp DNA circular BCT 05-SEP-2023 HDRNA_12_K01_bold_37467.current.gb:LOCUS CP133685 43849 bp DNA circular BCT 05-SEP-2023 HDRNA_12_K01_bold_37467.current.gb:LOCUS CP133686 6642 bp DNA circular BCT 05-SEP-2023 HDRNA_12_K01_bold_37467.current.gb:LOCUS CP133687 2241 bp DNA circular BCT 05-SEP-2023 HDRNA_16_K01_conservative_37834.current.gb:LOCUS CP133688 2626440 bp DNA circular BCT 05-SEP-2023 HDRNA_16_K01_conservative_37834.current.gb:LOCUS CP133689 24906 bp DNA circular BCT 05-SEP-2023 HDRNA_16_K01_conservative_37834.current.gb:LOCUS CP133690 20828 bp DNA circular BCT 05-SEP-2023 HDRNA_16_K01_conservative_37834.current.gb:LOCUS CP133691 4567 bp DNA circular BCT 05-SEP-2023 HDRNA_16_K01_conservative_37834.current.gb:LOCUS CP133692 2242 bp DNA circular BCT 05-SEP-2023 HDRNA_17_K01_conservative_37288.current.gb:LOCUS CP133693 2503072 bp DNA circular BCT 05-SEP-2023 HDRNA_17_K01_conservative_37288.current.gb:LOCUS CP133694 29861 bp DNA circular BCT 05-SEP-2023 HDRNA_17_K01_conservative_37288.current.gb:LOCUS CP133695 4439 bp DNA circular BCT 05-SEP-2023 HDRNA_19_K01_bold_37377.current.gb:LOCUS CP133696 2362062 bp DNA circular BCT 05-SEP-2023 HDRNA_19_K01_bold_37377.current.gb:LOCUS CP133697 55320 bp DNA circular BCT 05-SEP-2023 HDRNA_19_K01_bold_37377.current.gb:LOCUS CP133698 46464 bp DNA circular BCT 05-SEP-2023 HDRNA_19_K01_bold_37377.current.gb:LOCUS CP133699 11827 bp DNA circular BCT 05-SEP-2023 HDRNA_20_K01_conservative_43457.current.gb:LOCUS CP133700 2490778 bp DNA circular BCT 05-SEP-2023 HDRNA_20_K01_conservative_43457.current.gb:LOCUS CP133701 2241 bp DNA circular BCT 05-SEP-2023 ln -s /home/jhuang/Tools/spandx/ spandx nextflow run spandx/main.nf --fastq "trimmed_HDRNA_01/*_P_{1,2}.fastq.gz" --ref db/CP133676.fasta --annotation --database CP133676 -resume mv work work_CP133676 mv Outputs Outputs_CP133676 for fasta_file in CP133677 CP133678 CP133679 CP133680 CP133681 CP133682 CP133683 CP133684 CP133685 CP133686 CP133687 CP133688 CP133689 CP133690 CP133691 CP133692 CP133693 CP133694 CP133695 CP133696 CP133697 CP133698 CP133699 CP133700 CP133701; do echo "nextflow run spandx/main.nf --fastq "trimmed_HDRNA_01/*_P_{1,2}.fastq.gz" --ref db/${fasta_file}.fasta --annotation --database ${fasta_file} -resume" echo "mv work work_${fasta_file}" echo "mv Outputs Outputs_${fasta_file}" done for file in *.fastq.gz; do mv $file $(echo $file | cut -d'_' -f1)-$(echo $file | cut -d'_' -f1)-$(echo $file | cut -d'_' -f3)_$(echo $file | cut -d'_' -f6); done for file in *.fastq.gz; do mv $file $(echo $file | cut -d'_' -f3)_$(echo $file | cut -d'_' -f6); done for file in *.fastq.gz; do mv $file $(echo $file | cut -d'_' -f1)-$(echo $file | cut -d'_' -f2); done for file in *.fastq.gz; do mv $file $(echo $file | cut -d'-' -f1)_$(echo $file | cut -d'-' -f2); done Input read files could not be found. Have you included the read files in the current directory and do they have the correct naming? With the parameters specified, SPANDx is looking for reads named *_{1,2}.fastq.gz. To fix this error either rename your reads to match this formatting or specify the desired format when initializing SPANDx e.g. --fastq "*_{1,2}_sequence.fastq.gz" cd trimmed_HDRNA_01 nextflow run ../spandx/main.nf --ref ../db/CP133676.fasta --annotation --database CP133676 -resume mv Outputs Outputs_CP133676 cd .. cd trimmed_HDRNA_03 nextflow run ../spandx/main.nf --ref ../db/CP133677.fasta --annotation --database CP133677 -resume mv Outputs Outputs_CP133677 cd .. cd trimmed_HDRNA_06 nextflow run ../spandx/main.nf --ref ../db/CP133678.fasta --annotation --database CP133678 -resume mv work work_CP133678 mv Outputs Outputs_CP133678 nextflow run ../spandx/main.nf --ref ../db/CP133679.fasta --annotation --database CP133679 -resume mv work work_CP133679 mv Outputs Outputs_CP133679 cd .. cd trimmed_HDRNA_07 nextflow run ../spandx/main.nf --ref ../db/CP133680.fasta --annotation --database CP133680 -resume mv work work_CP133680 mv Outputs Outputs_CP133680 nextflow run ../spandx/main.nf --ref ../db/CP133681.fasta --annotation --database CP133681 -resume mv work work_CP133681 mv Outputs Outputs_CP133681 cd .. cd trimmed_HDRNA_08 nextflow run ../spandx/main.nf --ref ../db/CP133682.fasta --annotation --database CP133682 -resume mv work work_CP133682 mv Outputs Outputs_CP133682 nextflow run ../spandx/main.nf --ref ../db/CP133683.fasta --annotation --database CP133683 -resume mv work work_CP133683 mv Outputs Outputs_CP133683 cd .. cd trimmed_HDRNA_12 nextflow run ../spandx/main.nf --ref ../db/CP133684.fasta --annotation --database CP133684 -resume mv work work_CP133684 mv Outputs Outputs_CP133684 nextflow run ../spandx/main.nf --ref ../db/CP133685.fasta --annotation --database CP133685 -resume mv work work_CP133685 mv Outputs Outputs_CP133685 nextflow run ../spandx/main.nf --ref ../db/CP133686.fasta --annotation --database CP133686 -resume mv work work_CP133686 mv Outputs Outputs_CP133686 nextflow run ../spandx/main.nf --ref ../db/CP133687.fasta --annotation --database CP133687 -resume mv work work_CP133687 mv Outputs Outputs_CP133687 cd .. cd trimmed_HDRNA_16 nextflow run ../spandx/main.nf --ref ../db/CP133688.fasta --annotation --database CP133688 -resume mv work work_CP133688 mv Outputs Outputs_CP133688 nextflow run ../spandx/main.nf --ref ../db/CP133689.fasta --annotation --database CP133689 -resume mv work work_CP133689 mv Outputs Outputs_CP133689 nextflow run ../spandx/main.nf --ref ../db/CP133690.fasta --annotation --database CP133690 -resume mv work work_CP133690 mv Outputs Outputs_CP133690 nextflow run ../spandx/main.nf --ref ../db/CP133691.fasta --annotation --database CP133691 -resume mv work work_CP133691 mv Outputs Outputs_CP133691 nextflow run ../spandx/main.nf --ref ../db/CP133692.fasta --annotation --database CP133692 -resume mv work work_CP133692 mv Outputs Outputs_CP133692 cd .. cd trimmed_HDRNA_17 nextflow run ../spandx/main.nf --ref ../db/CP133693.fasta --annotation --database CP133693 -resume mv work work_CP133693 mv Outputs Outputs_CP133693 nextflow run ../spandx/main.nf --ref ../db/CP133694.fasta --annotation --database CP133694 -resume mv work work_CP133694 mv Outputs Outputs_CP133694 nextflow run ../spandx/main.nf --ref ../db/CP133695.fasta --annotation --database CP133695 -resume mv work work_CP133695 mv Outputs Outputs_CP133695 cd .. cd trimmed_HDRNA_19 nextflow run ../spandx/main.nf --ref ../db/CP133696.fasta --annotation --database CP133696 -resume mv work work_CP133696 mv Outputs Outputs_CP133696 nextflow run ../spandx/main.nf --ref ../db/CP133697.fasta --annotation --database CP133697 -resume mv work work_CP133697 mv Outputs Outputs_CP133697 nextflow run ../spandx/main.nf --ref ../db/CP133698.fasta --annotation --database CP133698 -resume mv work work_CP133698 mv Outputs Outputs_CP133698 nextflow run ../spandx/main.nf --ref ../db/CP133699.fasta --annotation --database CP133699 -resume mv work work_CP133699 mv Outputs Outputs_CP133699 cd .. cd trimmed_HDRNA_20 nextflow run ../spandx/main.nf --ref ../db/CP133700.fasta --annotation --database CP133700 -resume mv work work_CP133700 mv Outputs Outputs_CP133700 nextflow run ../spandx/main.nf --ref ../db/CP133701.fasta --annotation --database CP133701 -resume mv work work_CP133701 mv Outputs Outputs_CP133701 cd .. #------------------------- rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_01/Outputs_CP133676 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_03/Outputs_CP133677 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_06/Outputs_CP133678 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_06/Outputs_CP133679 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_07/Outputs_CP133680 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_07/Outputs_CP133681 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_08/Outputs_CP133682 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_08/Outputs_CP133683 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_12/Outputs_CP133684 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_12/Outputs_CP133685 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_12/Outputs_CP133686 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_12/Outputs_CP133687 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_16/Outputs_CP133688 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_16/Outputs_CP133689 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_16/Outputs_CP133690 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_16/Outputs_CP133691 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_16/Outputs_CP133692 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_17/Outputs_CP133693 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_17/Outputs_CP133694 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_17/Outputs_CP133695 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_19/Outputs_CP133696 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_19/Outputs_CP133697 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_19/Outputs_CP133698 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_19/Outputs_CP133699 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_20/Outputs_CP133700 . rsync -a -P jhuang@hamm:/home/jhuang/DATA/Data_Holger_S.epidermidis_short/trimmed_HDRNA_20/Outputs_CP133701 . cut -f2 -d$'\t' snippy.core.tab > f2 cut -f3 -d$'\t' snippy.core.tab > f3 cut -f4 -d$'\t' snippy.core.tab > f4 diff snippy/merged_snp.vcf.id variants/f2 5d5 < 138824 7d6 < 139197 9d7 < 139844 61d58 < 2475573 # -- merging vcf-files using bcftools -- results_HDRNA_01/snippy bcftools merge HDRNA_01_K01/HDRNA_01_K01.vcf.gz HDRNA_01_K02/HDRNA_01_K02.vcf.gz HDRNA_01_K03/HDRNA_01_K03.vcf.gz HDRNA_01_K04/HDRNA_01_K04.vcf.gz HDRNA_01_K05/HDRNA_01_K05.vcf.gz HDRNA_01_K06/HDRNA_01_K06.vcf.gz HDRNA_01_K07/HDRNA_01_K07.vcf.gz HDRNA_01_K08/HDRNA_01_K08.vcf.gz HDRNA_01_K09/HDRNA_01_K09.vcf.gz HDRNA_01_K10/HDRNA_01_K10.vcf.gz -o merged.vcf #bcftools index merged.vcf.gz cp merged.vcf merged_CP133676.vcf cd results_HDRNA_03/snippy bcftools merge HDRNA_03_K01/HDRNA_03_K01.vcf.gz HDRNA_03_K02/HDRNA_03_K02.vcf.gz HDRNA_03_K03/HDRNA_03_K03.vcf.gz HDRNA_03_K04/HDRNA_03_K04.vcf.gz HDRNA_03_K05/HDRNA_03_K05.vcf.gz HDRNA_03_K06/HDRNA_03_K06.vcf.gz HDRNA_03_K07/HDRNA_03_K07.vcf.gz HDRNA_03_K08/HDRNA_03_K08.vcf.gz HDRNA_03_K09/HDRNA_03_K09.vcf.gz HDRNA_03_K10/HDRNA_03_K10.vcf.gz -o merged.vcf cp merged.vcf merged_CP133677.vcf cd results_HDRNA_06/snippy bcftools merge HDRNA_06_K01/HDRNA_06_K01.vcf.gz HDRNA_06_K02/HDRNA_06_K02.vcf.gz HDRNA_06_K03/HDRNA_06_K03.vcf.gz HDRNA_06_K04/HDRNA_06_K04.vcf.gz HDRNA_06_K05/HDRNA_06_K05.vcf.gz HDRNA_06_K06/HDRNA_06_K06.vcf.gz HDRNA_06_K07/HDRNA_06_K07.vcf.gz HDRNA_06_K08/HDRNA_06_K08.vcf.gz HDRNA_06_K09/HDRNA_06_K09.vcf.gz HDRNA_06_K10/HDRNA_06_K10.vcf.gz -o merged.vcf #split merged.vcf to merged_CP133678.vcf and merged_CP133679.vcf cd results_HDRNA_07/snippy bcftools merge HDRNA_07_K01/HDRNA_07_K01.vcf.gz HDRNA_07_K02/HDRNA_07_K02.vcf.gz HDRNA_07_K03/HDRNA_07_K03.vcf.gz HDRNA_07_K04/HDRNA_07_K04.vcf.gz HDRNA_07_K05/HDRNA_07_K05.vcf.gz HDRNA_07_K06/HDRNA_07_K06.vcf.gz HDRNA_07_K07/HDRNA_07_K07.vcf.gz HDRNA_07_K08/HDRNA_07_K08.vcf.gz HDRNA_07_K09/HDRNA_07_K09.vcf.gz HDRNA_07_K10/HDRNA_07_K10.vcf.gz -o merged.vcf cp merged.vcf merged_CP133680.vcf #Note that merged_CP133681.vcf is empty. cd results_HDRNA_08/snippy bcftools merge HDRNA_08_K01/HDRNA_08_K01.vcf.gz HDRNA_08_K02/HDRNA_08_K02.vcf.gz HDRNA_08_K03/HDRNA_08_K03.vcf.gz HDRNA_08_K04/HDRNA_08_K04.vcf.gz HDRNA_08_K05/HDRNA_08_K05.vcf.gz HDRNA_08_K06/HDRNA_08_K06.vcf.gz HDRNA_08_K07/HDRNA_08_K07.vcf.gz HDRNA_08_K08/HDRNA_08_K08.vcf.gz HDRNA_08_K09/HDRNA_08_K09.vcf.gz HDRNA_08_K10/HDRNA_08_K10.vcf.gz -o merged.vcf #split merged.vcf to merged_CP133682.vcf and merged_CP133683.vcf. #----ERROR: IGNORING the record----> #CP133683 1718 . G A 155736 . QR=0;RO=0;ANN=A|intron_variant|MODIFIER|RGR12_11570|GENE_RGR12_11570|transcript|TRANSCRIPT_RGR12_11570|protein_coding|1/1|c.358-506C>T||||||WARNING_TRANSCRIPT_NO_START_CODON;DP=12890;AB=0;AO=3768;QA=121701;TYPE=snp GT:DP:RO:QR:AO:QA:GL ./.:.:.:.:.:.:. 1/1:5361:0:0:5347:173706:-15627.9,-1609.61,0 1/1:3750:0:0:3747:121191:-10903.3,-1127.96,0 ./.:.:.:.:.:.:. 1/1:3779:0:0:3768:121701:-10949.3,-1134.28,0 ./.:.:.:.:.:.:. ./.:.:.:.:.:.:. ./.:.:.:.:.:.:. ./.:.:.:.:.:.:. ./.:.:.:.:.:.:. cd results_HDRNA_12/snippy bcftools merge HDRNA_12_K01/HDRNA_12_K01.vcf.gz HDRNA_12_K02/HDRNA_12_K02.vcf.gz HDRNA_12_K03/HDRNA_12_K03.vcf.gz HDRNA_12_K04/HDRNA_12_K04.vcf.gz HDRNA_12_K05/HDRNA_12_K05.vcf.gz HDRNA_12_K06/HDRNA_12_K06.vcf.gz HDRNA_12_K07/HDRNA_12_K07.vcf.gz HDRNA_12_K08/HDRNA_12_K08.vcf.gz HDRNA_12_K09/HDRNA_12_K09.vcf.gz HDRNA_12_K10/HDRNA_12_K10.vcf.gz -o merged.vcf #split merged.vcf to merged_CP133684.vcf and merged_CP133685.vcf. #Note that merged_CP133686.vcf and merged_CP133687.vcf are empty. cd results_HDRNA_16/snippy bcftools merge HDRNA_16_K01/HDRNA_16_K01.vcf.gz HDRNA_16_K02/HDRNA_16_K02.vcf.gz HDRNA_16_K03/HDRNA_16_K03.vcf.gz HDRNA_16_K04/HDRNA_16_K04.vcf.gz HDRNA_16_K05/HDRNA_16_K05.vcf.gz HDRNA_16_K06/HDRNA_16_K06.vcf.gz HDRNA_16_K07/HDRNA_16_K07.vcf.gz HDRNA_16_K08/HDRNA_16_K08.vcf.gz HDRNA_16_K09/HDRNA_16_K09.vcf.gz HDRNA_16_K10/HDRNA_16_K10.vcf.gz -o merged.vcf cp merged.vcf merged_CP133688.vcf #Note that merged_CP133689.vcf - merged_CP133692.vcf are empty. cd results_HDRNA_17/snippy bcftools merge HDRNA_17_K01/HDRNA_17_K01.vcf.gz HDRNA_17_K02/HDRNA_17_K02.vcf.gz HDRNA_17_K03/HDRNA_17_K03.vcf.gz HDRNA_17_K04/HDRNA_17_K04.vcf.gz HDRNA_17_K05/HDRNA_17_K05.vcf.gz HDRNA_17_K06/HDRNA_17_K06.vcf.gz HDRNA_17_K07/HDRNA_17_K07.vcf.gz HDRNA_17_K08/HDRNA_17_K08.vcf.gz HDRNA_17_K09/HDRNA_17_K09.vcf.gz HDRNA_17_K10/HDRNA_17_K10.vcf.gz -o merged.vcf cp merged.vcf merged_CP133693.vcf #Note that merged_CP133694.vcf - merged_CP133695.vcf are empty. cd results_HDRNA_19/snippy bcftools merge HDRNA_19_K01/HDRNA_19_K01.vcf.gz HDRNA_19_K02/HDRNA_19_K02.vcf.gz HDRNA_19_K03/HDRNA_19_K03.vcf.gz HDRNA_19_K04/HDRNA_19_K04.vcf.gz HDRNA_19_K05/HDRNA_19_K05.vcf.gz HDRNA_19_K06/HDRNA_19_K06.vcf.gz HDRNA_19_K07/HDRNA_19_K07.vcf.gz HDRNA_19_K08/HDRNA_19_K08.vcf.gz HDRNA_19_K09/HDRNA_19_K09.vcf.gz HDRNA_19_K10/HDRNA_19_K10.vcf.gz -o merged.vcf cp merged.vcf merged_CP133696.vcf #Note that merged_CP133697.vcf - merged_CP133699.vcf are empty. cd results_HDRNA_20/snippy bcftools merge HDRNA_20_K01/HDRNA_20_K01.vcf.gz HDRNA_20_K02/HDRNA_20_K02.vcf.gz HDRNA_20_K03/HDRNA_20_K03.vcf.gz HDRNA_20_K04/HDRNA_20_K04.vcf.gz HDRNA_20_K05/HDRNA_20_K05.vcf.gz HDRNA_20_K06/HDRNA_20_K06.vcf.gz HDRNA_20_K07/HDRNA_20_K07.vcf.gz HDRNA_20_K08/HDRNA_20_K08.vcf.gz HDRNA_20_K09/HDRNA_20_K09.vcf.gz -o merged.vcf cp merged.vcf merged_CP133700.vcf #Note that merged_CP133701.vcf is empty. #Note not enough reads exist in 'HDRNA_20 S K10 365_BB84_S87_R1_001.fastq.gz' and 'HDRNA_20 S K10 365_BB84_S87_R2_001.fastq.gz'. python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_01/snippy/merged_CP133676.vcf Outputs_CP133676/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_01_not_in_vcf_file_output.txt HDRNA_01_not_in_txt_file_output.txt HDRNA_01_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_03/snippy/merged_CP133677.vcf Outputs_CP133677/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_03_not_in_vcf_file_output.txt HDRNA_03_not_in_txt_file_output.txt HDRNA_03_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_06/snippy/merged_CP133678.vcf Outputs_CP133678/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_06_1_not_in_vcf_file_output.txt HDRNA_06_1_not_in_txt_file_output.txt HDRNA_06_1_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_06/snippy/merged_CP133679.vcf Outputs_CP133679/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_06_2_not_in_vcf_file_output.txt HDRNA_06_2_not_in_txt_file_output.txt HDRNA_06_2_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_07/snippy/merged_CP133680.vcf Outputs_CP133680/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_07_not_in_vcf_file_output.txt HDRNA_07_not_in_txt_file_output.txt HDRNA_07_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_08/snippy/merged_CP133682.vcf Outputs_CP133682/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_08_not_in_vcf_file_output.txt HDRNA_08_not_in_txt_file_output.txt HDRNA_08_common_records_output.txt #ERROR: python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_08/snippy/merged_CP133683.vcf Outputs_CP133683/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_08_2_not_in_vcf_file_output.txt HDRNA_08_2_not_in_txt_file_output.txt HDRNA_08_2_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_12/snippy/merged_CP133684.vcf Outputs_CP133684/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_12_1_not_in_vcf_file_output.txt HDRNA_12_1_not_in_txt_file_output.txt HDRNA_12_1_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_12/snippy/merged_CP133685.vcf Outputs_CP133685/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_12_2_not_in_vcf_file_output.txt HDRNA_12_2_not_in_txt_file_output.txt HDRNA_12_2_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_16/snippy/merged_CP133688.vcf Outputs_CP133688/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_16_not_in_vcf_file_output.txt HDRNA_16_not_in_txt_file_output.txt HDRNA_16_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_17/snippy/merged_CP133693.vcf Outputs_CP133693/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_17_not_in_vcf_file_output.txt HDRNA_17_not_in_txt_file_output.txt HDRNA_17_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_19/snippy/merged_CP133696.vcf Outputs_CP133696/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_19_not_in_vcf_file_output.txt HDRNA_19_not_in_txt_file_output.txt HDRNA_19_common_records_output.txt python3 ~/Scripts/merge_snippy_and_spandx_results.py results_HDRNA_20/snippy/merged_CP133700.vcf Outputs_CP133700/Phylogeny_and_annotation/All_SNPs_indels_annotated.txt HDRNA_20_not_in_vcf_file_output.txt HDRNA_20_not_in_txt_file_output.txt HDRNA_20_common_records_output.txt mv HDRNA_01_common_records_output.txt _HDRNA_01.txt mv HDRNA_03_common_records_output.txt HDRNA_03.txt mv HDRNA_06_common_records_output.txt HDRNA_06.txt mv HDRNA_07_common_records_output.txt HDRNA_07.txt mv HDRNA_08_common_records_output.txt HDRNA_08.txt mv HDRNA_12_common_records_output.txt HDRNA_12.txt mv HDRNA_16_common_records_output.txt HDRNA_16.txt mv HDRNA_17_common_records_output.txt HDRNA_17.txt mv HDRNA_19_common_records_output.txt HDRNA_19.txt mv HDRNA_20_common_records_output.txt HDRNA_20.txt sed -i '1s/_trimmed_P//g' _HDRNA_01.txt sed -i '1s/_trimmed_P//g' HDRNA_03.txt HDRNA_06.txt HDRNA_07.txt HDRNA_08.txt HDRNA_12.txt HDRNA_16.txt HDRNA_17.txt HDRNA_19.txt HDRNA_20.txt # -- check if f3==f6 -- cut -f3 -d$'\t' HDRNA_17.txt > f3 cut -f6 -d$'\t' HDRNA_17.txt > f6 diff f3 f6 # -- check if f6==f7 in HDRNA_7.txt since they have the sample names -- cut HDRNA_07.txt -d$'\t' -f6 > f6 cut HDRNA_07.txt -d$'\t' -f7 > f7 diff d6 f7 --> delete the column HDRNA_07_K01-BB28 in variant_calling.xls. ~/Tools/csv2xls-0.4/csv_to_xls.py _HDRNA_01.txt HDRNA_03.txt HDRNA_06.txt HDRNA_07.txt HDRNA_08.txt HDRNA_12.txt HDRNA_16.txt HDRNA_17.txt HDRNA_19.txt HDRNA_20.txt -d$'\t' -o variant_calling.xls -
processing commands for presence phage HH1, SPbeta-like phage, phage related island
#makeblastdb -in HDRNA_K01.fna -dbtype nucl blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_01.blastn blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_01.blastn blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_03.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_03.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_06.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_06.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_07.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_07.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_08.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_08.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_12.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_12.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_16.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_16.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_17.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_17.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_19.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_19.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query MT880870.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880870_on_20.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query MT880871.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880871_on_20.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query MT880872.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > MT880872_on_20.blastn ΦSepi-HH1(MT880870): 34053 bp in ST2, ST83 (34053 bt) PI-Sepi-HH2(MT880871): not in ST2, but in ST290, ST297 and ST487 (36164 bt) ΦSPbeta-like(MT880872): in ST2 and ST22 (147057 bt) # In the new isolates, we have the MLST - ST130 ST224 ST23 ST35 ST487 --> ST5 ST640 ST69 ST86 ST87 --> HDRNA_17_K01 (ST69) contains MT880871! It agrees with the description in the Anna's paper, ST487 has PI-Sepi-HH2! #shovill/HDRNA_11_K01/contigs.fa sepidermidis 487 arcC(1) aroE(1) gtr(1) mutS(5) pyrR(2) tpiA(1) yqiL(1) # -- 01 -- 33342 34053 --> am Grenzen 33634 34132 --> am Grenzen 131829 147057 + 1 3810 --> am Grenzen 30857 34053 --> NA 33634 34132 --> NA 131829 147057 + 1 3807 --> NA NA NA NA 07: NA 08: NA 12: NA 16: NA 17: MT880871_on_17.blastn YES 19: NA 20: NA shovill/HDRNA_11_K01/contigs.fa sepidermidis 487 arcC(1) aroE(1) gtr(1) mutS(5) pyrR(2) tpiA(1) yqiL(1) a Biofilm formation compared to S. epidermidis 1457: –, ⬍30%; ⫹, 30% to 59%; ⫹⫹, 60% to 84%; ⫹⫹⫹, ⱖ85%. b ST, sequence type determined by MLST. c GC, genetic cluster based on Bayesian analysis of population structure (BAPS) of MLST. d ND, not determined. In columns 2 to 5, ⫹ indicates presence and ⫺ indicates absence. ND, not determined or defined. '+' indicates presence and '-' indicates absence. SPbeta-like staphylococcus phage (NC_029119.1, 86% sequence identity)[47] and will here be referred to as ΦSPbeta-like, (GenBank accession number MT880872) the remaining two regions did not correspond to any previously described Staphylococcus phage[48] ΦSepi-HH1 (MT880870): 34053 bp phage-related island PI-Sepi-HH2 (MT880871). MT880870.1: Staphylococcus phage PhiSepi-HH1, complete genome MT880871.1: Staphylococcus phage PI-Sepi-HH2, complete genome: 36164 bp MT880872.1: Staphylococcus phage PhiSepi-HH3, complete genome -
processing commands for the other genes from Gene_List.pptx
# AND "Staphylococcus epidermidis"[porgn:__txid1282] mv HDRNA_16_K01_conservative_37834.current.gb ../gbk mv ./HDRNA_17_K01_conservative_37288.current.gb ../gbk mv ./HDRNA_03_K01_bold_bandage_26831.current.gb ../gbk mv ./HDRNA_06_K01_conservative_27645.current.gb ../gbk mv ./HDRNA_12_K01_bold_37467.current.gb ../gbk mv ./HDRNA_01_K01_conservative_23197.current.gb ../gbk mv ./HDRNA_07_K01_conservative_27169.current.gb ../gbk mv ./HDRNA_08_K01_conservative_32455.current.gb ../gbk mv ./HDRNA_19_K01_bold_37377.current.gb ../gbk mv ./HDRNA_20_K01_conservative_43457.current.gb ../gbk (agrC) AND "Staphylococcus" https://www.ncbi.nlm.nih.gov/nuccore/OQ828637.1 gltA agrC gene 2184799..2186091 /gene="agrC" /locus_tag="SAR2125" CDS 2184799..2186091 /gene="agrC" /locus_tag="SAR2125" /note="Signal dectecting component of the agr autoinducer peptide-quorum sensing system. Two-component regulatory system family, sensor kinase protein. Similar to Staphylococcus aureus accessory gene regulator C AgrC TR:Q53644 (EMBL:X52543) (423 aa) fasta scores: E(): 1.7e-101, 75.23% id in 424 aa, and to Staphylococcus epidermidis histidine kinase AgrC TR:O68159 (EMBL:AF012132) (429 aa) fasta scores: E(): 6.3e-76, 54.93% id in 426 aa" /codon_start=1 /transl_table=11 /product="autoinducer sensor protein" /protein_id="CAG41106.1" /db_xref="EnsemblGenomes-Gn:SAR2125" /db_xref="EnsemblGenomes-Tr:CAG41106" /translation="MEALNDYNYVLFVIVQVSLMFFISAFISGIRYKKSDYIYIIGIV LSSVYFFDKIRSISLVVITIFIIIFLYFKIRLYSVFLVMVTQIILYCANFVYIIIFSY IITISHSVFIVLPIFLVVYVSISYALAYILNRILKRINGTYLSLNKKFLTVITIVIVI TFSLLFAYSQIDASDASTIKQYSLLFLGIIILLSILIFIYSQFTLKEMKYKRNQEEIE TYYEYTLKIEAINNEMRKFRHDYVNILTTLSEYIREDDMTGLRDYFNKNIVPMKDNLQ MNALKLNGIENLKVREIKGLLTAKILRAQEMNIPISIEIPDEVTRINLNMIDLSRSIG IILDNAIEASSEIDDPIIRVAFIESENSVTFIVMNKCADDIPRIHELFQESFSTKGEG RGLGLSTLKEIADNADNVLLDTIIENGFFIQKVEIINN" MEALNDYNYVLFVIVQVSLMFFISAFISGIRYKKSDYIYIIGIVLSSVYFFDKIRSISLVVITIFIIIFLYFKIRLYSVFLVMVTQIILYCANFVYIIIFSYIITISHSVFIVLPIFLVVYVSISYALAYILNRILKRINGTYLSLNKKFLTVITIVIVITFSLLFAYSQIDASDASTIKQYSLLFLGIIILLSILIFIYSQFTLKEMKYKRNQEEIETYYEYTLKIEAINNEMRKFRHDYVNILTTLSEYIREDDMTGLRDYFNKNIVPMKDNLQMNALKLNGIENLKVREIKGLLTAKILRAQEMNIPISIEIPDEVTRINLNMIDLSRSIGIILDNAIEASSEIDDPIIRVAFIESENSVTFIVMNKCADDIPRIHELFQESFSTKGEGRGLGLSTLKEIADNADNVLLDTIIENGFFIQKVEIINN Staphylococcus_aureus_MRSA252 samtools faidx Staphylococcus_aureus_MRSA252.fasta "gi|49240382|emb|BX571856.1|":2184799-2186091 (yycG) AND "Staphylococcus" gene 25617..27443 /gene="yycG" /locus_tag="SAR0019" /gene_synonym="vicK" CDS 25617..27443 /gene="yycG" /locus_tag="SAR0019" /gene_synonym="vicK" /note="Two-component regulatory system family, sensor kinase protein. Previously sequenced as Staphylococcus aureus two-component sensor histidine kinase YycG TR:Q9XCM6 (EMBL:AF136709) (608 aa) fasta scores: E(): 5.5e-214, 99.836% id in 608 aa. Similar to Bacillus subtilis probable two-component sensor histidine kinase YycG TR:Q45614 (EMBL:D78193) (611 aa) fasta scores: E(): 2e-98, 46.217% id in 608 aa" /codon_start=1 /transl_table=11 /product="sensor kinase protein" /protein_id="CAG39047.1" /db_xref="EnsemblGenomes-Gn:SAR0019" /db_xref="EnsemblGenomes-Tr:CAG39047" /db_xref="GOA:Q6GKS6" /db_xref="InterPro:IPR000014" /db_xref="InterPro:IPR000700" /db_xref="InterPro:IPR003594" /db_xref="InterPro:IPR003660" /db_xref="InterPro:IPR003661" /db_xref="InterPro:IPR004358" /db_xref="InterPro:IPR005467" /db_xref="InterPro:IPR029151" /db_xref="UniProtKB/Swiss-Prot:Q6GKS6" /translation="MKWLKQLQSLHTKLVIVYVLLIIIGMQIIGLYFTNNLEKELLDN FKKNITQYAKQLEISIEKVYDEKGSVNAQKDIQNLLSEYANRQEIGEIRFIDKDQIII ATTKQSNRSLINQKANDSSVQKALSLGQSNDHLILKDYGGGKDRVWVYNIPVKVDKKV IGNIYIESKINDVYNQLNNINQIFIVGTAISLLITVILGFFIARTITKPITDMRNQTV EMSRGNYTQRVKIYGNDEIGELALAFNNLSKRVQEAQANTESEKRRLDSVITHMSDGI IATDRRGRIRIVNDMALKMLGMAKEDIIGYYMLSVLSLEDEFKLEEIQENNDSFLLDL NEEEGLIARVNFSTIVQETGFVTGYIAVLHDVTEQQQVERERREFVANVSHELRTPLT SMNSYIEALEEGAWKDEELAPQFLSVTREETERMIRLVNDLLQLSKMDNESDQINKEI IDFNMFINKIINRHEMSTKDTTFIRDIPKKTIFTEFDPDKMTQVFDNVITNAMKYSRG DKRVEFHVKQNPLYNRMTIRIKDNGIGIPINKVDKIFDRFYRVDKARTRKMGGTGLGL AISKEIVEAHNGRIWANSVEGQGTSIFITLPCEVIEDGDWDE" samtools faidx Staphylococcus_aureus_MRSA252.fasta "gi|49240382|emb|BX571856.1|":25617-27443 psmß1: https://www.ncbi.nlm.nih.gov/nuccore/380448412 hlb: https://www.ncbi.nlm.nih.gov/nuccore/KC242859.1 atlE tagB gene 694999..696102 /gene="tagB" /locus_tag="SAR0649" CDS 694999..696102 /gene="tagB" /locus_tag="SAR0649" /note="Similar to Bacillus subtilis teichoic acid biosynthesis protein B TagB SW:TAGB_BACSU (P27621) (381 aa) fasta scores: E(): 4.3e-26, 31.302% id in 361 aa, and to Lactococcus lactis teichoic acid biosynthesis protein B TagB TR:Q9CH14 (EMBL:AE006327) (371 aa) fasta scores: E(): 2.8e-06, 24.834% id in 302 aa. Lack of similarity at the N-terminus in comparison to other orthologues" /codon_start=1 /transl_table=11 /product="teichoic acid biosynthesis protein" /protein_id="CAG39666.1" /db_xref="EnsemblGenomes-Gn:SAR0649" /db_xref="EnsemblGenomes-Tr:CAG39666" /translation="MNVLIKKFYHLVVRILSKMITPQVIDKPHIVFMMTFPEDIKPII KALNNSLYQKTVLTTPKQAPYLSELSDDVNVIEMTNRTLVKQIKALKSAQMIIIDNYY LLLGGYNKTSNQHIVQTWHASGALKNFGLTDHQVDVSDKAMVQQYRKVYQATDFYLVG CEQMSQCFKQSLGATEEQMLYFGLPRINKYYTADRATVKAELKDKYGITNKLALYVPT YREDKADNRAIDKAYFEKCLPGYTLINKLHPSIEHSDIDDVSSIDTSILMLMSDIIIS DYSSLPIEASLLDIPTIFYVYDEGTYDKVRGLNQFYKAIPDSYKVYTEEDLIMTIQEK EHLLSPLFKDWHKYNTDKSLHQLTEYIDKMVTK" samtools faidx Staphylococcus_aureus_MRSA252.fasta "gi|49240382|emb|BX571856.1|":694999-696102 > tagB.fasta gene 2137..2586 /gene="capC" CDS 2137..2586 /gene="capC" /codon_start=1 /transl_table=11 /product="CapC" /protein_id="BAB13485.1" /translation="MFGSDLYIALILGVLLSLIFAEKTGIVPAGLVVPGYLGLVFNQP VFILLVLLVSLLTYVIVKYGLSKFMILYGRRKFAAMLITGIVLKIAFDFLYPIVPFEI AEFRGIGIIVPGLIANTIQKQGLTITFGSTLLLSGATFAIMFVYYLI" samtools faidx capBCA_ywtC.fasta "gi|10119860|dbj|AB039950.1|":2137-2586 > capC.fasta gene 2783..3256 /gene="sepA" CDS 2783..3256 /gene="sepA" /function="multidrug resistance" /experiment="experimental evidence, no additional details recorded" /note="similar to BAB43260.1 of S. aureus N315" /codon_start=1 /transl_table=11 /product="SepA" /protein_id="BAB83937.1" /translation="MIVNYLKHKFYNLLTTMIVLFIFVLSGAIFLTFLGFGLYGLSRI LIYFRLGDFTYNRSMYDNLLYYGSYIIFGYFIIFAVEHLMDYFRKMLPENAYFRGATF HLISYTVATTLFYFIIHLNYVYINIDFWVIMVIIGFLYVCKLQFYPESKNLNNRK" samtools faidx ORF123_sepA_ORF5.fasta "gi|18250967|dbj|AB078343.1|":2783-3256 > sepA.fasta gene 1427047..1429569 /gene="mprF" /locus_tag="SAR1372" /gene_synonym="fmtC" CDS 1427047..1429569 /gene="mprF" /locus_tag="SAR1372" /gene_synonym="fmtC" /note="Similar to Staphylococcus aureus putative membrane protein MprF TR:AAK58115 (EMBL:AF145699) (840 aa) fasta scores: E(): 0, 96.190% id in 840 aa. Similar to Staphylococcus xylosus putative membrane protein MprF TR:AAK58113 (EMBL:AF145698) (841 aa) fasta scores: E(): 5.4e-208, 62.530% id in 838 aa. Mutations in the CDS have reduced resistance to human defensins and evasion of neutrophil killing" /codon_start=1 /transl_table=11 /product="putative membrane protein" /protein_id="CAG40370.1" /db_xref="EnsemblGenomes-Gn:SAR1372" /db_xref="EnsemblGenomes-Tr:CAG40370" /db_xref="GOA:Q6GH45" /db_xref="InterPro:IPR016181" /db_xref="InterPro:IPR022791" /db_xref="InterPro:IPR024320" /db_xref="UniProtKB/Swiss-Prot:Q6GH45" /translation="MNQEVKNKIFSILKITFATALFIFVVITLYRELSGINFKDTLVE FSKINRMSLVLLFIGGGASLVILSMYDVILSRALKMDISLGKVLRVSYIINALNAIVG FGGFIGAGVRAMVYKNYTHDKKKLVHFISLILISMLTGLSLLSLLIVFHVFDASLILN KITWVRWVLYAVSLFLPLFIIYSMVRPPDKNNRYVGLYCTLVSCVEWLAAAVVLYFCG VIVDVHVSFMSFIAIFIIAALSGLVSFIPGGFGAFDLVVLLGFKTLGVPEEKVLLMLL LYRFAYYFVPVIIALILSSFEFGTSAKKYIEGSKYFIPAKDVTSFLMSYQKDIIAKIP SLSLAILVFFTSMIFFVNNLTIVYDALYDGNHLTYYLLLAIHTSACLLLLLNVVGIYK QSRRAIIYAMISIILIIVATLFTYASYILITWLVIIFALLIVAFRRARRLKRPIRMRN LVAMLLFSIFILYINHIFIAGTFYALDVYTIEMHTSVLKYYFWITILIIAIIVGAIAW LFDYQFSKVRISSNIEECEEIIDQYGGNYLSHLIYSGDKQFFTNEDKNAFLMYRYKAS SLVVLGDPIGDENAFDELLEAFYNYAEYLGYDVIFYQVTDQHMPLYHNFGNQFFKLGE EAIIDLTQFSTSGKKRRGFRATLNKFDELNISFEIIEPPFSTEFINELQHVSDLWLDN RQEMHFSVGQFNETYLSKAPIGVMRNENNEVIAFCSLMPTYFNDAISVDLIRWLPELD LPLMDGLYLHMLLWSKEQGYTKFNMGMATLSNVGQLHYSYLRERLAGRVFEHFNGLYR FQGLRRYKSKYNPNWEPRFLVYRKDNSLWESLSKVMRVIRHK" samtools faidx Staphylococcus_aureus_MRSA252.fasta "gi|49240382|emb|BX571856.1|":1427047-1429569 > fmtC.fasta gene 1825..2523 /gene="sceD" CDS 1825..2523 /gene="sceD" /codon_start=1 /transl_table=11 /product="SceD precursor" /protein_id="AAB94657.1" /translation="MKKLLVASSASAALFAVGVGANAHAAEDNNVNQDQLAQTALNNT QQLNDAPVQEGAYNIAFDNSGYNFNFNSDGTNWSWSYNADSSAQQAPAQSTTQEQAPA AQQAPAQSTTQEQAPAAQQAPAQEQTQQPAQQPAQQQTQQPAQQSADSGSNVQVNDHL KAIAQRESGGDIHAINSSSGAAGKYQFLQTTWDSVAPAEYQGKPASEAPEAVQDAAAQ KLYDTAGPSQWVTA" sig_peptide 1825..1899 /gene="sceD" mat_peptide 1900..2520 /gene="sceD" /product="SceD" samtools faidx sceDAE.fasta "gi|6707000|gb|AF109218.1|":1825-2523 > sceD.fasta gene 759097..759894 /locus_tag="SERP_RS03845" /old_locus_tag="SE0760" /old_locus_tag="SERP0760" CDS 759097..759894 /locus_tag="SERP_RS03845" /old_locus_tag="SE0760" /old_locus_tag="SERP0760" /inference="COORDINATES: similar to AA sequence:RefSeq:WP_001830106.1" /note="Derived by automated computational analysis using gene prediction method: Protein Homology." /codon_start=1 /transl_table=11 /product="VOC family protein" /protein_id="WP_002446237.1" /db_xref="GI:488376852" /translation="MFHNKNANFVNGVTINIKDKETIKTFYENVLGFNLINESKTAVQ FEVGDSNQFITFIEIQNGREPLMSEAGLFHIGILLPTLTTLADLLVHLSDFEVPVNGG QQSVATCLFIEDPEGNAIKFYVDRETESWIDEKEGRIRMDIAPINVPRLLQNVSHTQW QGIPDETKLGSLHIKSIRISDVKSYYLHYFGLEESAYMDDYSLFLSSNDYYNHLAVNQ WLSATKRVDNEHTYGLAMIDFHYPKTTHKNLKGPDGIYFRFNRIKEV" samtools faidx Staphylococcus_epidermidis_RP62A.fasta "gi|57865352|ref|NC_002976.3|":759097-759894 > SE0760.fasta gene 3091..8793 /gene="esp" /allele="1" CDS 3091..8793 /gene="esp" /allele="1" /note="enterococcal surface protein; ORF3" /codon_start=1 /transl_table=11 /product="Esp" /protein_id="AAQ84025.1" /translation="MVSKNNKRVFLEKTKKRVLKYSIKKLSVGVASVLVGVGLVLGTT ELVQAQDEISPSTPLETAISSVQVGDKVASGNTFQENPGYTKNYNFSDLQFSPQELTG DTLKGNTIGFEVYGKHNIAASTKNWEIRLQLDERLAKYVEKIQVDPKKGIGSSRRTFV RINDSLGRPTNIWKVNYIRASDGLFAGAETTDTQTAPNGVITFEKSLDEIFKEIGIDN LKTDRLMYRIYLVSHQDDDKIVPGIDSTGYFLTDSDDFYNSLDVSENNPDQFKHGSVS AKYEEPNTQTKDGSGSTGANGAIILDHKLTKNYNFSYSASAKGTPWYANYKIDERLVP YVAGIQMHMVQADKVTYDVSFESGKKVADLAIERRKDHENYGVGSITDNDLTKLIDFA NASPRPVVIRYVLQLTKPLDEILEDMKATAQVEENKPFGEDFIFDSWLSDTNKKLIQN TYGTGYYYLQDIDGDGNPDDKEESGDTNPYIGKPELEEVYDVDTTVKGKVFIHELAGT GHKAQLVDKEGTVLAEKTIAPNEKDGAPISDTVEFEFTGVDSSKLIAKDELKIQIVSP GFDKPEEGSTVIKESPKAVDKQTVVVGFKPDAKESIRNNKNLPEDAEYSWKTEPDTSN VTDSTKGIVTVKIGNRTFDVDVEFAVKASQAMENDATYVPITTTPETTVQSGKPTFDK PDVPLAKDAFSILDVYNKDFGNASVDANTGIVTFTPAKGVGESEPITGTIPIKIVYQD GSVGTTDLAVTVSKDIYENPGENIPAGYHKVTFTAGEGTSIESGTTVFAVKDGVSLPE DKLPVLKAKDGYTDAKWPEEATQPITADDTEFVSSATKLDDIIENPGENIPAGYHKVT FTAGEGTSIESGTTVFAVKDGVSLPEDKLPVLKAKDGYTDAKWPEEATQPITADDTEF VSSATKLDDIIENPGENIPAGYHKVTFTAGEGTSIESGTTVFAVKDGVSLPEDKLPVL KAKDGYTDAKWPEEATQPITADDTEFVSSATKLDDIIENPGENIPAGYHKVIFTAGEG TSIESGTTVFAVKDGVSLPEDKLPVLKAKDGYTDAKWPEEATQPITADDTEFVSSATK LDDIIENPGENIPAGYHKVIFTAGEGTSIESGTTVFAVKDGVSLPEDKLPVLKAKDGY TDAKWPEEATQPITADDTEFVSSATKLDDKSDADKYNPEGQKVTTELNKEPDASEGIK NKEDLPKDTKYTWKEKVDVSAAGNKKGTVVVTYSDGSSDEVEVDVTVTDNRSDADKYE PTVEGEKVEVGGTVDLTDNVTNLPTLPEGTTVTDVTPDGTIDTNTPGNYEGVIEVTYP DGTKDTVKVPVEVTDNRSDADKYTPMVEGEKVEVGGTVDLTDNVTNLPTLPEGTTVTD VTPDGTIDTNTPGNYEGVIEVTYPDGTKDTVKVPVEVTDNRSDADKYTPMVEGEKVEV GGTVDLTDNVTNLPTLPEGTTVTDVTPGGTIDTNTPGNYEGVIEVTYPDGTKDTVKVP VEVTDNRSDADKYEPTVEGEKVEVGGTVDLTDNVTNLPTLPEGTTVTDVTPGGTIDTN TPGNYEGVIEVTYPDGTKDTVKVPVEVTDNRSDADKYTPMVEGEKVEVGGTVDLTDNV TNLPTLPEGTTVTDVTPDGTIDTNTPGNYEGVIEVTYPDGTKDTVKVPVEVTDNRSDA DKYNPEGQKVTTDLNKEPDASEGIKNKEDLPKGTTYTWKEKVDVSTAGNKKGIVVVTY PDGSKEEVEVTISVEDKKAPNKPQVDPITEGDQIVTGKTEPNAEVTVTLPNGSQYHGT ADKNGNFTVKVPKLEAGTKVIVTATDESGNTSEPTNVVVSSNEKDSEKAVSKDNKTDN QGSKQNTNRGKSSPQKQSSKAYPKTGEIDSNIFTISGGLILLGTLGLLGYKNRKKENE " samtools faidx Enterococcus_faecium_isolate_E300_pathogenicity_island.fasta "gi|34980227|gb|AY322150.1|":3091-8793 > esp.fasta mv ~/Downloads/sequence\ \(1\).fasta ecpA.fasta mv ~/Downloads/sequence\ \(1\).fasta sdrG.fasta HH1-HP1 HH1-HP2 HH1-TreR sdrG.fasta gltA.fasta agrC.fasta yycG.fasta atlE.fasta psm-beta1.fasta gene 366..500 /gene="psm beta1" CDS 366..500 /gene="psm beta1" /note="hemolytic protein; pfam05480" /codon_start=1 /transl_table=11 /product="Psm beta1" /protein_id="AFD54319.1" /translation="MEGLFNAIKDTVTAAINNDGAKLGTSIVSIVENGVGLLGKLFGF " gene 557..691 /gene="psm beta2" CDS 557..691 /gene="psm beta2" /note="hemolytic protein; pfam05480" /codon_start=1 /transl_table=11 /product="Psm beta2" /protein_id="AFD54320.1" /translation="MTGLAEAIANTVQAAQQHDSVKLGTSIVDIVANGVGLLGKLFGF " samtools faidx psm-beta.fasta "gi|380448412|gb|JQ066320.1|":366-500 > psm-beta1.fasta hlb.fasta samtools faidx hlb_.fasta "gi|441494790|gb|KC242859.1|":16-1008 > hlb.fasta tagB.fasta capC.fasta sepA.fasta fmtC.fasta sceD.fasta SE0760.fasta esp.fasta samtools faidx ecpA_.fasta "gi|354620065|gb|JN051494.1|":248-835 > ecpA.fasta ecpA.fasta #TODO TOMORROW: add the gene names in the fasta and merge them in a file using the following command line, make once blastn, oder make gene by gene. It is maybe clearer! find the last 3 gene sequences. At first, send the results except for gene arrangements! #cat sdrG.fasta gltA.fasta agrC.fasta yycG.fasta atlE.fasta psm-beta1.fasta hlb.fasta tagB.fasta capC.fasta sepA.fasta fmtC.fasta sceD.fasta SE0760.fasta esp.fasta ecpA.fasta > 15genes.fasta #makeblastdb -in HDRNA_K01.fna -dbtype nucl # -- sdrG (repeated) -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query sdrG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sdrG_on_20.blastn # -- gltA -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query gltA.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > gltA_on_20.blastn #-->None # -- agrC -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query agrC.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > agrC_on_20.blastn #-->None # -- yycG (1827 nt) -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query yycG.fasta -evalue 1e-50 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > yycG_on_20.blastn #-- atlE.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query atlE.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > atlE_on_20.blastn #-- psm-beta1.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query psm-beta1.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > psm-beta1_on_20.blastn #-- hlb.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query hlb.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > hlb_on_20.blastn #-- tagB.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query tagB.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > tagB_on_20.blastn #-- capC.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query capC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > capC_on_20.blastn #-- sepA.fasta -- blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query sepA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sepA_on_20.blastn fmtC.fasta blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query fmtC.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > fmtC_on_20.blastn sceD.fasta blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query sceD.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > sceD_on_20.blastn SE0760.fasta blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query SE0760.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > SE0760_on_20.blastn esp.fasta blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query esp.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > esp_on_20.blastn ecpA.fasta blastn -db HDRNA_01_K01_conservative_23197.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_01.blastn blastn -db HDRNA_03_K01_bold_bandage_26831.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_03.blastn blastn -db HDRNA_06_K01_conservative_27645.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_06.blastn blastn -db HDRNA_07_K01_conservative_27169.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_07.blastn blastn -db HDRNA_08_K01_conservative_32455.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_08.blastn blastn -db HDRNA_12_K01_bold_37467.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_12.blastn blastn -db HDRNA_16_K01_conservative_37834.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_16.blastn blastn -db HDRNA_17_K01_conservative_37288.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_17.blastn blastn -db HDRNA_19_K01_bold_37377.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_19.blastn blastn -db HDRNA_20_K01_conservative_43457.current.gb_converted.fna -query ecpA.fasta -evalue 1e-10 -num_threads 15 -outfmt 6 -strand both -max_target_seqs 1 > ecpA_on_20.blastn 12+15+3-1=29 I only found ebpS instead of ebp. #(fumC) AND "complete cds" gyrB: https://www.ncbi.nlm.nih.gov/nuccore/MG995415.1 Mycobacterium tuberculosis strain UKR100 GyrB (gyrB) gene, complete cds fumC: https://www.ncbi.nlm.nih.gov/nuccore/3153897 gltA: icd: -
Bioinformatics Tools:
Artemis: A genome browser and annotation tool that can generate similar kinds of schematic representations. * GenomeDiagram from Biopython: A toolkit within Biopython that allows for the creation of high-quality genomic graphics. #--> https://biopython-tutorial.readthedocs.io/en/latest/notebooks/17%20-%20Graphics%20including%20GenomeDiagram.html IGV (Integrative Genomics Viewer): While primarily a genome browser, IGV can be used to generate snapshots that display genomic regions and variations. General Graphic Design Software: Adobe Illustrator: A popular graphic design tool used to create precise and detailed scientific figures. Inkscape: A free and open-source vector graphics editor that can be used to create diagrams like the one you've shown. Specialized Genomic Visualization Tools: SnapGene or Geneious: While mainly used for plasmid mapping and editing, these tools also offer features to create genomic maps and diagrams. MEGA (Molecular Evolutionary Genetics Analysis): Primarily used for evolutionary analyses, MEGA can also produce graphical representations of genes and genomes. Online Tools: PlasMapper: Used primarily for plasmid maps but can be adapted for smaller genomic regions. ApE (A plasmid Editor): Another tool for visualizing plasmid maps that might be used to create simplified genomic diagrams. Without specific information, it's hard to pinpoint exactly which one was used for your image, but it's likely that the original creator used one of the bioinformatics tools or a combination of general graphic design software to illustrate the genomic rearrangements. If you're looking to create similar images, you might want to explore some of these options to see which one best suits your needs.
点赞本文的读者
还没有人对此文章表态
本文有评论
没有评论
看文章,发评论,不要沉默
最受欢迎文章
- Motif Discovery in Biological Sequences: A Comparison of MEME and HOMER
- Calling peaks using findPeaks of HOMER
- Kraken2 Installation and Usage Guide
- Why Do Significant Gene Lists Change After Adding Additional Conditions in Differential Gene Expression Analysis?
- Should the inputs for GSVA be normalized or raw?
- PiCRUST2 Pipeline for Functional Prediction and Pathway Analysis in Metagenomics
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- pheatmap vs heatmap.2
- Setup conda environments
- Guide to Submitting Data to GEO (Gene Expression Omnibus)
最新文章
- Risks of Rebooting into Rescue Mode
- 足突(Podosome)、胞外囊泡(Extracellular Vesicle)与基质金属蛋白酶(MMPs)综合解析
- NCBI BioSample Submission Strategy for PJI and Nasal Microbiota Study
- Human RNA-seq processing for Data_Ben_RNAseq_2025
最多评论文章
- Updating Human Gene Identifiers using Ensembl BioMart: A Step-by-Step Guide
- The top 10 genes
- Retrieving KEGG Genes Using Bioservices in Python
推荐相似文章
Data_Tam_DNAseq_2025 NACHREICHEN_2
DNAseq 2025_AYE and DNAseq 2025_adeABadeIJ_adeIJK_CM1_CM2
Using vrap and bacto to process Acinetobacter baumannii isolates